Our lab has developed a number of protocols for gene targeting and allele replacement in yeast. Our current focus is to facilitate manipulation of whole genomes by recombination using targetting DNAs produced by PCR. We have thus far developed several PCR-based gene manipulation techniques including gene disruption, allele replacement, and epitope tagging with cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP). Each of these methods integrates DNA into the genome using a selectable marker that can later be removed via direct repeat recombination, thus keeping that marker "open" for later gene manipulations. In addition, we are also developing methods to facilitate screening of arrayed yeast libraries, including 96-well format transformation protocols and kar1-mediated plasmoduction as an alternate, more rapid method of transferring hundreds of plasmids in a simple replica-plating experiment.
|
Adaptamers:
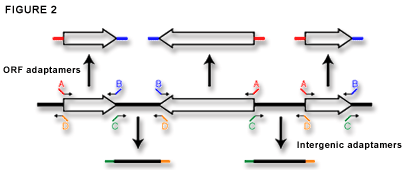
There are two genome-wide sets of adaptamers that are commercially available from Research Genetics. The complete set of ORF adaptamers contain fusion tags that we refer to as A and B (Figure 2). These are uniformly oriented so that all forward ORF adaptamers bind starting at the ATG of the gene and contain the A fusion sequence. Every reverse adaptamer binds starting at the ORF stop codon and contains the B fusion sequence. Furthermore, the A sequence maintains a reading frame so that gene fusions can be easily produced. Figure 2 also shows the adaptamer set designed to amplify all non-ORF or intergenic regions in the yeast genome. We use this set for our PCR-based gene disruption protocol described below. These adaptamers are uniformly oriented from left to right on the map of each chromosome so that all forward adaptamers contain the C fusion sequence and all reverse primers contain the D fusion sequence. |
|
|
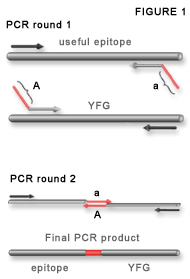
Figure 1.
Adaptamer-mediated PCR fusion. Adaptamers are illustrated as chimeric primers containing a homologous sequence (parallel to the template DNA) and a nonhomologous fusion tag (diagonal) labelled A and a. The fusions tags are reverse and complementary so they anneal in the second round of PCR to make a fusion DNA. The final product contains each domain separated by the 20mer fusion sequence.
Figure 2.
|
![]()
Our gene disruption protocol uses the set of intergenic adaptamers to amplify long regions of homology flanking the gene to be disrupted. The use of long homology regions increases the gene targeting efficiency. The C and D adaptamer tags are then used to fuse the flanking DNAs to a dominant selectable marker gene in a second round of PCR. The selectable marker is also designed to be recyclable by direct repreat recombination. Thus the sets of reagents used to perform a gene disruption are generic and can be used over and over to perform multiple gene disruptions in the same strain.
The gene disruption efficiency and a detailed protocol can be found in the following publications:
-
Reid, R.J.D., Sunjevaric, I.Keddache, M. and Rothstein, R.
Efficient PCR-based gene disruption in Saccharomyces strains using intergenic primers.
Yeast 19: 319-328, 2002.
- Reid, R. Lisby, M. and Rothstein, R. Cloning-free genome alterations in Saccharomyces cerevisiae using adaptamer-mediated PCR. In: Methods in Enzymology, Vol. 350., G. R. Fink and C. Guthrie, Eds., Academic Press, New York, pp. 258-277, 2002.
In addition the following plasmid maps and sequence files are available:
-
Plasmid pWJ1077 contains the K. lactis URA3 gene with flanking 143 bp direct repeats.
Sequence file [Genbank]* [FastA]
Primer map [PDF]
Restriction site map [PDF] -
Plasmid pWJ716 contains the K. lactis URA3 gene without direct repeats.
Sequence file [Genbank]* [FastA]
Primer map [PDF]
Restriction site map [PDF]
Intergenic Primer Search Page:
Intergenic primer sets used to perform gene disruptions are available from Invitrogen/Research Genetics. We have organized the database of primer sets so that is searchable by gene name in order to identify the primers to knock out any particular gene.
Use the following link to access our intergenic primer search page:
Site Directed Mutagenesis / RFLP Calculator:
The yeast mutation calculator allows you to change a codon to introduce a single amino acid change and then see all the sequence permutations in the 15 nucleotides surrounding that change. A list of restriction sites is generated for each sequence permutation to help define useful restriction polymorphisms for the mutated sequence. In addition, the product of the codon frequencies for the sequence are listed for comparison to the wild-type sequence.
Use the following link to access this tool:
Fusing novel epitopes to a gene of interest provides a number of useful tools to understand protein function. We use adaptamer-mediated PCR to integrate CFP and YFP fusions directly into the genome. After selection for the CFP or YFP integrants, the selectable marker is removed by direct repeat recombination so that only the epitope is integrated into the coding region of interest. This means that N-terminal, C-terminal and even internal integrations of an epitope are possible, and in each case, transcriptional control is maintained by the endogenous promoter. Recent results using Rad52-YFP and a detailed protocol can be found in the following publications:
-
Lisby, M., Rothstein, R. and Mortensen, U.H.
Rad52 forms DNA repair and recombination centers during S phase.
Proc. Natl. Acad. Sci. USA 98: 8276-8282, 2001.
- Reid, R. Lisby, M. and Rothstein, R. Cloning-free genome alterations in Saccharomyces cerevisiae using adaptamer-mediated PCR. In: Methods in Enzymology, Vol. 350., G. R. Fink and C. Guthrie, Eds., Academic Press, New York, pp. 258-277, 2002.
![]()
kar-mediated plasmid transfer:
We have developed and successfully used a method for the efficient transfer of a single plasmid into multiple recipient strains or many different plasmids into a single recipient. The method is based on yeast mating and uses a particular mutant allele, kar1∆15, that is proficient in mating but defective in nuclear fusion. This property of kar1∆15 allows a plasmid to transfer from one (donor) strain to another (recipient) strain without mixing of the all the chromosomes from the two nuclei. This method is described in the following manuscript:
- Georgieva, B. and Rothstein, R. kar-mediated plasmid transfer between yeast strains, an alternative to traditional transformation methods. In: Methods in Enzymology, Vol. 350., G. R. Fink and C. Guthrie, Eds., Academic Press, New York, pp. 278-289, 2002.
96-well Yeast LiOAc Transformation:
We have also been using LiOAc transformations in a 96-well format to transfer plasmids into 96 different strains at once. Follow the link below to read our protocol.